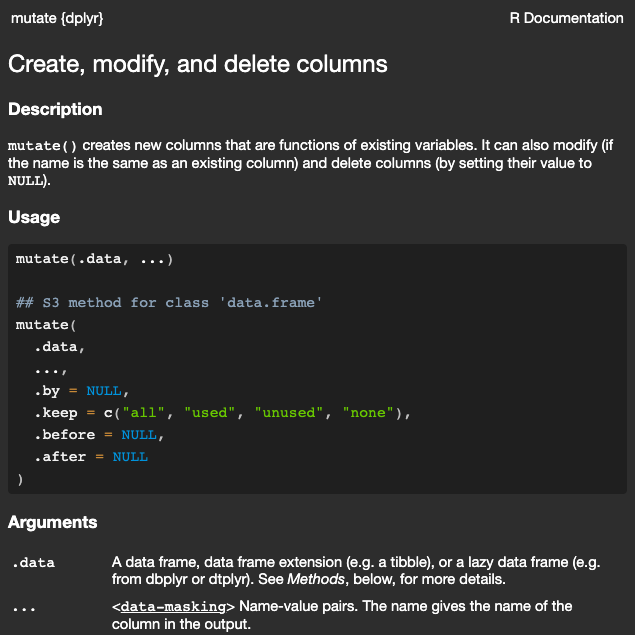
set.seed(5000)
df <- tibble(
a = rnorm(5),
b = rnorm(5),
c = rnorm(5),
d = rnorm(5)
)
df
df |> mutate(
a = (a - min(a, na.rm = TRUE)) /
(max(a, na.rm = TRUE) - min(a, na.rm = TRUE)),
b = (b - min(b, na.rm = TRUE)) /
(max(b, na.rm = TRUE) - min(a, na.rm = TRUE)),
c = (c - min(c, na.rm = TRUE)) /
(max(c, na.rm = TRUE) - min(c, na.rm = TRUE)),
d = (d - min(d, na.rm = TRUE)) /
(max(d, na.rm = TRUE) - min(d, na.rm = TRUE))
)- 1
-
set.seed()allows you to reproduce the same numbers from any random number-generating process (i.e.rnorm())