- 1
- Wrapping an entire object assignment in parentheses simultaneously defines the object and shows you what it represents.
[1] 1 2Manipulating and Summarizing Data
CS&SS 508 • Lecture 4
21 October 2025
Victoria Sass
R
Tools like Excel or Google Sheets let you manipulate spreadsheets using functions.
Today, we’ll use R to manipulate data more transparently and reproducibly.
R?Under the hood, R stores different types of data in different ways.
4.0 is a number, and that "Vic" is not a number.So what exactly are the common data types, and how do we know what R is doing?
logical)factor)Date, POSIXct, POSIXt)integer, double)NA, NaN, Inf)character)c(FALSE, TRUE, TRUE)factor(c("red", "blue"))as_Date(c("2018-10-04"))c(1, 10*3, 4, -3.14)c(NA, NA, NA, NaN, NaN, NA)c("red", "blue", "blue")The simplest data type is a Boolean, or binary, variable: TRUE or FALSE1.
More often than not our data don’t actually have a variable with this data type, but they are definitely created and evaluated in the data manipulation and summarizing process.
Logical operators refer to base functions which allow us to test if a condition is present between two objects.
For example, we may test
Naturally, these types of expressions produce a binary outcome of T or F which enables us to transform our data in a variety of ways!
R==:!=:>, >=, <, <=:%in%:&:|:!:xor():TRUE becomes FALSE, vice versa)Be careful using == with numbers:
[1] 1 2Similarly mysterious, missing values (NA) represent the unknown. Almost anything conditional involving NAs will also be unknown:
Let’s create two objects, A and B
Comparisons:
Combinations:
B < 10 | > 20). The code won’t technically error but it won’t evaluate the way you expect it to. Read more about the confusing logic behind this here. In essence the truncated second part of this conditional statement (> 20) will evaluate to TRUE since any numeric that isn’t 0 for a logical operator is coerced to TRUE. Therefore this statement will actually always evaluate to TRUE and will return all elements of B instead of the ones that meet your specified condition.
[1] FALSE TRUE TRUE
[1] TRUE FALSE TRUE
[1] TRUE FALSE TRUEany():all():|; it’ll return TRUE if there are any TRUE’s in x&; it’ll return TRUE only if all values of x are TRUE’sif_else()
If you want to use one value when a condition is TRUE and another value when it’s FALSE.
case_when()
A very useful extension of if_else() for multiple conditions1.
.default if you want to create a “default”/catch all value.
NA and everything.
[1] "-ve" "-ve" "-ve" "0" "+ve" "+ve" "+ve" "???"dplyrToday, we’ll use tools from the dplyr package to manipulate data!
ggplot2, dplyr is part of the Tidyverse, and included in the tidyverse package.To demonstrate data transformations we’re going to use the nycflights13 dataset, which you’ll need to download and load into R
nycflights13 includes five data frames1:, some of which contain missing data (NA):
dplyr BasicsAll dplyr functions have the following in common:
Each function operates either on rows, columns, groups, or entire tables.
To save the transformations you’ve made to a data frame you’ll need to save the output to a new object.
filter()We often get big datasets, and we only want some of the entries. We can subset rows using filter().
== not = to test the logical condition.
delay_2hr is an object in our environment which contains rows corresponding to flights that experienced at least a 2 hour delay.
# A tibble: 9,723 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 848 1835 853 1001 1950
2 2013 1 1 957 733 144 1056 853
3 2013 1 1 1114 900 134 1447 1222
4 2013 1 1 1540 1338 122 2020 1825
5 2013 1 1 1815 1325 290 2120 1542
6 2013 1 1 1842 1422 260 1958 1535
7 2013 1 1 1856 1645 131 2212 2005
8 2013 1 1 1934 1725 129 2126 1855
9 2013 1 1 1938 1703 155 2109 1823
10 2013 1 1 1942 1705 157 2124 1830
# ℹ 9,713 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>select()What if we want to keep every observation, but only use certain variables? Use select()!
We can select columns by name:
- before a variable name or a vector of variables to drop them from the data (i.e. select(-c(year, month, day))).
# A tibble: 336,776 × 3
year month day
<int> <int> <int>
1 2013 1 1
2 2013 1 1
3 2013 1 1
4 2013 1 1
5 2013 1 1
6 2013 1 1
7 2013 1 1
8 2013 1 1
9 2013 1 1
10 2013 1 1
# ℹ 336,766 more rowsselect()What if we want to keep every observation, but only use certain variables? Use select()!
We can select columns between variables (inclusive):
select()What if we want to keep every observation, but only use certain variables? Use select()!
We can select columns based on a condition:
select() including starts_with(), ends_with(), contains() and num_range(). Read more about these and more here.
# A tibble: 336,776 × 4
carrier tailnum origin dest
<chr> <chr> <chr> <chr>
1 UA N14228 EWR IAH
2 UA N24211 LGA IAH
3 AA N619AA JFK MIA
4 B6 N804JB JFK BQN
5 DL N668DN LGA ATL
6 UA N39463 EWR ORD
7 B6 N516JB EWR FLL
8 EV N829AS LGA IAD
9 B6 N593JB JFK MCO
10 AA N3ALAA LGA ORD
# ℹ 336,766 more rowsdistinct()You may want to find the unique combinations of variables in a dataset. Use distinct()
distinct() drops variables!By default, distinct() drops unused variables. If you don’t want to drop them, add the argument .keep_all = TRUE:
distinct() will find the first occurrence of a unique row in the dataset and discard the rest. Use count() if you’re looking for the number of occurrences.
# A tibble: 224 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 214 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>count()sort = TRUE arranges them in descending order of number of occurrences.
# A tibble: 224 × 3
origin dest n
<chr> <chr> <int>
1 JFK LAX 11262
2 LGA ATL 10263
3 LGA ORD 8857
4 JFK SFO 8204
5 LGA CLT 6168
6 EWR ORD 6100
7 JFK BOS 5898
8 LGA MIA 5781
9 JFK MCO 5464
10 EWR BOS 5327
# ℹ 214 more rowsarrange()Sometimes it’s useful to sort rows in your data, in ascending (low to high) or descending (high to low) order. We do that with arrange().
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>arrange()To sort in descending order, using desc() within arrange()
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 9 641 900 1301 1242 1530
2 2013 6 15 1432 1935 1137 1607 2120
3 2013 1 10 1121 1635 1126 1239 1810
4 2013 9 20 1139 1845 1014 1457 2210
5 2013 7 22 845 1600 1005 1044 1815
6 2013 4 10 1100 1900 960 1342 2211
7 2013 3 17 2321 810 911 135 1020
8 2013 6 27 959 1900 899 1236 2226
9 2013 7 22 2257 759 898 121 1026
10 2013 12 5 756 1700 896 1058 2020
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>rename()You may receive data with unintuitive variable names. Change them using rename().
rename(new_name = old_name) is the format. Reminder to use janitor::clean_names() if you want to automate this process for a lot of variables.
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tail_num <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>Variable Syntax
I recommend against using spaces in a name! It makes things really hard sometimes!!
mutate()You can add new columns to a data frame using mutate().
mutate() adds new columns on the right hand side of your dataset, which makes it difficult to see if anything happened. You can use the .before argument to specify which numeric index (or variable name) to move the newly created variable to. .after is an alternative argument for this.
# A tibble: 336,776 × 21
gain speed year month day dep_time sched_dep_time dep_delay arr_time
<dbl> <dbl> <int> <int> <int> <int> <int> <dbl> <int>
1 -9 370. 2013 1 1 517 515 2 830
2 -16 374. 2013 1 1 533 529 4 850
3 -31 408. 2013 1 1 542 540 2 923
4 17 517. 2013 1 1 544 545 -1 1004
5 19 394. 2013 1 1 554 600 -6 812
6 -16 288. 2013 1 1 554 558 -4 740
7 -24 404. 2013 1 1 555 600 -5 913
8 11 259. 2013 1 1 557 600 -3 709
9 5 405. 2013 1 1 557 600 -3 838
10 -10 319. 2013 1 1 558 600 -2 753
# ℹ 336,766 more rows
# ℹ 12 more variables: sched_arr_time <int>, arr_delay <dbl>, carrier <chr>,
# flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>,
# distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>mutate()You can specify which columns to keep with the .keep argument:
flights |>
mutate(
gain = dep_delay - arr_delay,
hours = air_time / 60,
gain_per_hour = gain / hours,
.keep = "used"
)"used" retains only the variables used to create the new variables, which is useful for checking your work. Other options include: "all" (default, returns all columns), "unused" (columns not used to create new columns) and "none" (only grouping variables and columns created by mutate are retained).
# A tibble: 336,776 × 6
dep_delay arr_delay air_time gain hours gain_per_hour
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2 11 227 -9 3.78 -2.38
2 4 20 227 -16 3.78 -4.23
3 2 33 160 -31 2.67 -11.6
4 -1 -18 183 17 3.05 5.57
5 -6 -25 116 19 1.93 9.83
6 -4 12 150 -16 2.5 -6.4
7 -5 19 158 -24 2.63 -9.11
8 -3 -14 53 11 0.883 12.5
9 -3 -8 140 5 2.33 2.14
10 -2 8 138 -10 2.3 -4.35
# ℹ 336,766 more rowsrelocate()You might want to collect related variables together or move important variables to the front. Use relocate()!
relocate() moves variables to the front but you can also specify where to put them using the .before and .after arguments, just like in mutate().
# A tibble: 336,776 × 19
time_hour air_time year month day dep_time sched_dep_time
<dttm> <dbl> <int> <int> <int> <int> <int>
1 2013-01-01 05:00:00 227 2013 1 1 517 515
2 2013-01-01 05:00:00 227 2013 1 1 533 529
3 2013-01-01 05:00:00 160 2013 1 1 542 540
4 2013-01-01 05:00:00 183 2013 1 1 544 545
5 2013-01-01 06:00:00 116 2013 1 1 554 600
6 2013-01-01 05:00:00 150 2013 1 1 554 558
7 2013-01-01 06:00:00 158 2013 1 1 555 600
8 2013-01-01 06:00:00 53 2013 1 1 557 600
9 2013-01-01 06:00:00 140 2013 1 1 557 600
10 2013-01-01 06:00:00 138 2013 1 1 558 600
# ℹ 336,766 more rows
# ℹ 12 more variables: dep_delay <dbl>, arr_time <int>, sched_arr_time <int>,
# arr_delay <dbl>, carrier <chr>, flight <int>, tailnum <chr>, origin <chr>,
# dest <chr>, distance <dbl>, hour <dbl>, minute <dbl>group_by()If you want to analyze your data by specific groupings, use group_by():
group_by() doesn’t change the data but you’ll notice that the output indicates that it is “grouped by” month (Groups: month [12]). This means subsequent operations will now work “by month”.
# A tibble: 336,776 × 19
# Groups: month [12]
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>summarize()summarize() calculates summaries of variables in your data:
You can use any function inside summarize() that aggregates multiple values into a single value (like sd(), mean(), or max()).
summarize() ExampleLet’s see what this looks like in our flights dataset:
summarize() ExampleLet’s see what this looks like in our flights dataset:
What if we want to summarize data by our groups? Use group_by() and summarize()
Because we did group_by() with month, then used summarize(), we get one row per value of month!
You can create any number of summaries in a single call to summarize().
n() returns the number of rows in each group.
# A tibble: 12 × 3
month delay n
<int> <dbl> <int>
1 1 10.0 27004
2 2 10.8 24951
3 3 13.2 28834
4 4 13.9 28330
5 5 13.0 28796
6 6 20.8 28243
7 7 21.7 29425
8 8 12.6 29327
9 9 6.72 27574
10 10 6.24 28889
11 11 5.44 27268
12 12 16.6 28135# A tibble: 336,776 × 19
# Groups: year, month, day [365]
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>Summary & Grouping Behavior
When you summarize a tibble grouped by more than one variable, each summary peels off the last group. You can change the default behavior by setting the .groups argument to a different value, e.g., "drop" to drop all grouping or "keep" to preserve the same groups. The default is "drop_last" if all groups have 1 row and keep otherwise (it’s recommended to use reframe() if this is the case, which is a more general version of summarize() that allows for an arbitrary number of rows per group and drops all grouping variables after execution).
ungroup()# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>.by.by works with all verbs and has the advantage that you don’t need to use the .groups argument to suppress the grouping message or ungroup() when you’re done.
# A tibble: 12 × 3
month delay n
<int> <dbl> <int>
1 1 10.0 27004
2 10 6.24 28889
3 11 5.44 27268
4 12 16.6 28135
5 2 10.8 24951
6 3 13.2 28834
7 4 13.9 28330
8 5 13.0 28796
9 6 20.8 28243
10 7 21.7 29425
11 8 12.6 29327
12 9 6.72 27574slice_*There are five handy functions that allow you extract specific rows within each group:
df |> slice_head(n = 1) takes the first row from each group.df |> slice_tail(n = 1) takes the last row in each group.df |> slice_min(x, n = 1) takes the row with the smallest value of column x.df |> slice_max(x, n = 1) takes the row with the largest value of column x.df |> slice_sample(n = 1) takes one random row.Let’s find the flights that are most delayed upon arrival at each destination.
slice_*n to select more than one row, or instead of n, you can use prop to select a proportion (between 0 and 1) of the rows in each group.
# A tibble: 108 × 19
# Groups: dest [105]
dest arr_delay year month day dep_time sched_dep_time dep_delay arr_time
<chr> <dbl> <int> <int> <int> <int> <int> <dbl> <int>
1 ABQ 153 2013 7 22 2145 2007 98 132
2 ACK 221 2013 7 23 1139 800 219 1250
3 ALB 328 2013 1 25 123 2000 323 229
4 ANC 39 2013 8 17 1740 1625 75 2042
5 ATL 895 2013 7 22 2257 759 898 121
6 AUS 349 2013 7 10 2056 1505 351 2347
7 AVL 228 2013 8 13 1156 832 204 1417
8 BDL 266 2013 2 21 1728 1316 252 1839
9 BGR 238 2013 12 1 1504 1056 248 1628
10 BHM 291 2013 4 10 25 1900 325 136
# ℹ 98 more rows
# ℹ 10 more variables: sched_arr_time <int>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, air_time <dbl>, distance <dbl>, hour <dbl>,
# minute <dbl>, time_hour <dttm>gapminder1
year (ascending order) and population (descending order) and save that over (i.e. overwrite) the object created for answer 1. Print the first 6 rows.ggplot, create a line plot showing life expectancy over time by country. Make the plot visually appealing!Question 1:
subset_gapminder <- gapminder |>
filter(country %in% c("China","India","United States"), year > 1980 ) |>
select(country, year, pop, lifeExp)
subset_gapminderfilter() by separating them with commas
# A tibble: 18 × 4
country year pop lifeExp
<fct> <int> <int> <dbl>
1 China 1982 1000281000 65.5
2 China 1987 1084035000 67.3
3 China 1992 1164970000 68.7
4 China 1997 1230075000 70.4
5 China 2002 1280400000 72.0
6 China 2007 1318683096 73.0
7 India 1982 708000000 56.6
8 India 1987 788000000 58.6
9 India 1992 872000000 60.2
10 India 1997 959000000 61.8
11 India 2002 1034172547 62.9
12 India 2007 1110396331 64.7
13 United States 1982 232187835 74.6
14 United States 1987 242803533 75.0
15 United States 1992 256894189 76.1
16 United States 1997 272911760 76.8
17 United States 2002 287675526 77.3
18 United States 2007 301139947 78.2Question 2:
[1] 18 4Rows: 18
Columns: 4
$ country <fct> "China", "China", "China", "China", "China", "China", "India",…
$ year <int> 1982, 1987, 1992, 1997, 2002, 2007, 1982, 1987, 1992, 1997, 20…
$ pop <int> 1000281000, 1084035000, 1164970000, 1230075000, 1280400000, 13…
$ lifeExp <dbl> 65.525, 67.274, 68.690, 70.426, 72.028, 72.961, 56.596, 58.553…[1] 18 4Question 3:
arrange() is to sort in ascending order
Question 4:
# A tibble: 18 × 5
country year pop lifeExp pop_billions
<fct> <int> <int> <dbl> <dbl>
1 China 1982 1000281000 65.5 1.00
2 India 1982 708000000 56.6 0.708
3 United States 1982 232187835 74.6 0.232
4 China 1987 1084035000 67.3 1.08
5 India 1987 788000000 58.6 0.788
6 United States 1987 242803533 75.0 0.243
7 China 1992 1164970000 68.7 1.16
8 India 1992 872000000 60.2 0.872
9 United States 1992 256894189 76.1 0.257
10 China 1997 1230075000 70.4 1.23
11 India 1997 959000000 61.8 0.959
12 United States 1997 272911760 76.8 0.273
13 China 2002 1280400000 72.0 1.28
14 India 2002 1034172547 62.9 1.03
15 United States 2002 287675526 77.3 0.288
16 China 2007 1318683096 73.0 1.32
17 India 2007 1110396331 64.7 1.11
18 United States 2007 301139947 78.2 0.301Question 5:
ungroup() is called). Learn more about this new feature here)
# A tibble: 6 × 2
year TotalPop_Billions
<int> <dbl>
1 1982 1.94
2 1987 2.11
3 1992 2.29
4 1997 2.46
5 2002 2.60
6 2007 2.73Question 6:
library(ggplot2)
library(ggthemes)
library(geomtextpath)
ggplot(subset_gapminder,
aes(year, lifeExp, color = country)) +
geom_point() +
geom_textpath(aes(label = country),
show.legend = FALSE) +
labs(title = "Life Expectancy (1982-2007)","China, India, and United States",
x = "Year",
y = "Life Expectancy (years)") +
scale_x_continuous(breaks = c(1982, 1987, 1992, 1997, 2002, 2007)) +
ylim(c(50, 80)) +
theme_tufte(base_size = 20)year to the x-axis, lifeExp to the y-axis, and country to color
geom_textpath() from the geomtextpath package to make nice labelled lines (specify mapping of country to the label)
ggthemes package and increase text size throughout plot
In practice, we often collect data from different sources. To analyze the data, we usually must first combine (merge) them.
For example, imagine you would like to study county-level patterns with respect to age and grocery spending. However, you can only find,
Merge the data!!
To do this we’ll be using the various join functions from the dplyr package.
We need to think about the following when we want to merge data frames A and B:
Keys are the way that two datasets are connected to one another. The two types of keys are:
Let’s look at our data to gain a better sense of what this all means.
airlines records two pieces of data about each airline: its carrier code and its full name. You can identify an airline with its two letter carrier code, making carrier the primary key.
glimpse on any dataset to see a transposed version of the data, making it possible to see all available column names, types, and a preview of as many values will fit on your current screen.
Rows: 16
Columns: 2
$ carrier <chr> "9E", "AA", "AS", "B6", "DL", "EV", "F9", "FL", "HA", "MQ", "O…
$ name <chr> "Endeavor Air Inc.", "American Airlines Inc.", "Alaska Airline…airports records data about each airport. You can identify each airport by its three letter airport code, making faa the primary key.
Rows: 1,458
Columns: 8
$ faa <chr> "04G", "06A", "06C", "06N", "09J", "0A9", "0G6", "0G7", "0P2", "…
$ name <chr> "Lansdowne Airport", "Moton Field Municipal Airport", "Schaumbur…
$ lat <dbl> 41.13047, 32.46057, 41.98934, 41.43191, 31.07447, 36.37122, 41.4…
$ lon <dbl> -80.61958, -85.68003, -88.10124, -74.39156, -81.42778, -82.17342…
$ alt <dbl> 1044, 264, 801, 523, 11, 1593, 730, 492, 1000, 108, 409, 875, 10…
$ tz <dbl> -5, -6, -6, -5, -5, -5, -5, -5, -5, -8, -5, -6, -5, -5, -5, -5, …
$ dst <chr> "A", "A", "A", "A", "A", "A", "A", "A", "U", "A", "A", "U", "A",…
$ tzone <chr> "America/New_York", "America/Chicago", "America/Chicago", "Ameri…planes records data about each plane. You can identify a plane by its tail number, making tailnum the primary key.
Rows: 3,322
Columns: 9
$ tailnum <chr> "N10156", "N102UW", "N103US", "N104UW", "N10575", "N105UW…
$ year <int> 2004, 1998, 1999, 1999, 2002, 1999, 1999, 1999, 1999, 199…
$ type <chr> "Fixed wing multi engine", "Fixed wing multi engine", "Fi…
$ manufacturer <chr> "EMBRAER", "AIRBUS INDUSTRIE", "AIRBUS INDUSTRIE", "AIRBU…
$ model <chr> "EMB-145XR", "A320-214", "A320-214", "A320-214", "EMB-145…
$ engines <int> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, …
$ seats <int> 55, 182, 182, 182, 55, 182, 182, 182, 182, 182, 55, 55, 5…
$ speed <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ engine <chr> "Turbo-fan", "Turbo-fan", "Turbo-fan", "Turbo-fan", "Turb…weather records data about the weather at the origin airports. You can identify each observation by the combination of location and time, making origin and time_hour the compound primary key.
Rows: 26,115
Columns: 15
$ origin <chr> "EWR", "EWR", "EWR", "EWR", "EWR", "EWR", "EWR", "EWR", "EW…
$ year <int> 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013,…
$ month <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ day <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ hour <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 15, 16, 17, 18, …
$ temp <dbl> 39.02, 39.02, 39.02, 39.92, 39.02, 37.94, 39.02, 39.92, 39.…
$ dewp <dbl> 26.06, 26.96, 28.04, 28.04, 28.04, 28.04, 28.04, 28.04, 28.…
$ humid <dbl> 59.37, 61.63, 64.43, 62.21, 64.43, 67.21, 64.43, 62.21, 62.…
$ wind_dir <dbl> 270, 250, 240, 250, 260, 240, 240, 250, 260, 260, 260, 330,…
$ wind_speed <dbl> 10.35702, 8.05546, 11.50780, 12.65858, 12.65858, 11.50780, …
$ wind_gust <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 20.…
$ precip <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ pressure <dbl> 1012.0, 1012.3, 1012.5, 1012.2, 1011.9, 1012.4, 1012.2, 101…
$ visib <dbl> 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,…
$ time_hour <dttm> 2013-01-01 01:00:00, 2013-01-01 02:00:00, 2013-01-01 03:00…flights has three variables (time_hour, flight, carrier) that uniquely identify an observation. More significantly, however, it contains foreign keys that correspond to the primary keys of the other datasets.
Rows: 336,776
Columns: 19
$ year <int> 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2…
$ month <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
$ day <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
$ dep_time <int> 517, 533, 542, 544, 554, 554, 555, 557, 557, 558, 558, …
$ sched_dep_time <int> 515, 529, 540, 545, 600, 558, 600, 600, 600, 600, 600, …
$ dep_delay <dbl> 2, 4, 2, -1, -6, -4, -5, -3, -3, -2, -2, -2, -2, -2, -1…
$ arr_time <int> 830, 850, 923, 1004, 812, 740, 913, 709, 838, 753, 849,…
$ sched_arr_time <int> 819, 830, 850, 1022, 837, 728, 854, 723, 846, 745, 851,…
$ arr_delay <dbl> 11, 20, 33, -18, -25, 12, 19, -14, -8, 8, -2, -3, 7, -1…
$ carrier <chr> "UA", "UA", "AA", "B6", "DL", "UA", "B6", "EV", "B6", "…
$ flight <int> 1545, 1714, 1141, 725, 461, 1696, 507, 5708, 79, 301, 4…
$ tailnum <chr> "N14228", "N24211", "N619AA", "N804JB", "N668DN", "N394…
$ origin <chr> "EWR", "LGA", "JFK", "JFK", "LGA", "EWR", "EWR", "LGA",…
$ dest <chr> "IAH", "IAH", "MIA", "BQN", "ATL", "ORD", "FLL", "IAD",…
$ air_time <dbl> 227, 227, 160, 183, 116, 150, 158, 53, 140, 138, 149, 1…
$ distance <dbl> 1400, 1416, 1089, 1576, 762, 719, 1065, 229, 944, 733, …
$ hour <dbl> 5, 5, 5, 5, 6, 5, 6, 6, 6, 6, 6, 6, 6, 6, 6, 5, 6, 6, 6…
$ minute <dbl> 15, 29, 40, 45, 0, 58, 0, 0, 0, 0, 0, 0, 0, 0, 0, 59, 0…
$ time_hour <dttm> 2013-01-01 05:00:00, 2013-01-01 05:00:00, 2013-01-01 0…These are variables in a dataset that refer to a primary key in another dataset. Note: pinkish shading indicates the primary key for that particular dataset.
flights$origin –> airports$faaflights$dest –> airports$faaflights$origin-flights$time_hour –> weather$origin-weather$time_hour.flights$tailnum –> planes$tailnumflights$carrier –> airlines$carrierA nice feature of these data are that the primary and foreign keys have the same name and almost every variable name used across multiple tables has the same meaning.1 This isn’t always the case!2
It is good practice to make sure your primary keys actually uniquely identify an observation and that they don’t have any missing values.
# A tibble: 0 × 2
# ℹ 2 variables: tailnum <chr>, n <int>Sometimes you’ll want to create an index of your observations to serve as a surrogate key because the compound primary key is not particularly easy to reference.
For example, our flights dataset has three variables that uniquely identify each observation: time_hour, carrier, flight.
row_number() simply specifies the row number of the data frame.
# A tibble: 336,776 × 20
id year month day dep_time sched_dep_time dep_delay arr_time
<int> <int> <int> <int> <int> <int> <dbl> <int>
1 1 2013 1 1 517 515 2 830
2 2 2013 1 1 533 529 4 850
3 3 2013 1 1 542 540 2 923
4 4 2013 1 1 544 545 -1 1004
5 5 2013 1 1 554 600 -6 812
6 6 2013 1 1 554 558 -4 740
7 7 2013 1 1 555 600 -5 913
8 8 2013 1 1 557 600 -3 709
9 9 2013 1 1 557 600 -3 838
10 10 2013 1 1 558 600 -2 753
# ℹ 336,766 more rows
# ℹ 12 more variables: sched_arr_time <int>, arr_delay <dbl>, carrier <chr>,
# flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>,
# distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>All join functions have the same basic interface: they take a pair of data frames and return one data frame.
The order of the rows and columns is primarily going to be determined by the first data frame.
dplyr has two types of joins: mutating and filtering.
Add new variables to one data frame from matching observations from another data frame.
left_join()right_join()inner_join()full_join()Filter observations from one data frame based on whether or not they match an observation in another data frame.
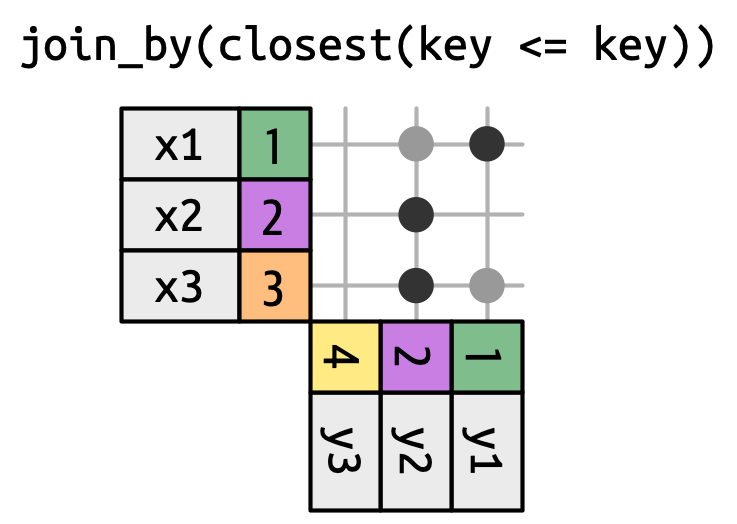
semi_join()anti-join()Mutating Joins
left_join()The most common type of join Appends columns from y to x by the rows in x
(NA added if there is nothing from y) Natural join: when all variables that appear in both datasets are used as the join key.
If the join_by() argument is not specified, left_join() will automatically join by all columns that have names and values in common.
left_join in nycflights13With only the pertinent variables from the flights dataset, we can see how a left_join works with the airlines dataset.
Joining with `by = join_by(carrier)`# A tibble: 336,776 × 7
year time_hour origin dest tailnum carrier name
<int> <dttm> <chr> <chr> <chr> <chr> <chr>
1 2013 2013-01-01 05:00:00 EWR IAH N14228 UA United Air Lines Inc.
2 2013 2013-01-01 05:00:00 LGA IAH N24211 UA United Air Lines Inc.
3 2013 2013-01-01 05:00:00 JFK MIA N619AA AA American Airlines Inc.
4 2013 2013-01-01 05:00:00 JFK BQN N804JB B6 JetBlue Airways
5 2013 2013-01-01 06:00:00 LGA ATL N668DN DL Delta Air Lines Inc.
6 2013 2013-01-01 05:00:00 EWR ORD N39463 UA United Air Lines Inc.
7 2013 2013-01-01 06:00:00 EWR FLL N516JB B6 JetBlue Airways
8 2013 2013-01-01 06:00:00 LGA IAD N829AS EV ExpressJet Airlines I…
9 2013 2013-01-01 06:00:00 JFK MCO N593JB B6 JetBlue Airways
10 2013 2013-01-01 06:00:00 LGA ORD N3ALAA AA American Airlines Inc.
# ℹ 336,766 more rowsplanes dataset has variables tailnum, year, type, manufacturer, model, engines, seats, speed, and engine
Joining with `by = join_by(year, tailnum)`# A tibble: 336,776 × 13
year time_hour origin dest tailnum carrier type manufacturer
<int> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
1 2013 2013-01-01 05:00:00 EWR IAH N14228 UA <NA> <NA>
2 2013 2013-01-01 05:00:00 LGA IAH N24211 UA <NA> <NA>
3 2013 2013-01-01 05:00:00 JFK MIA N619AA AA <NA> <NA>
4 2013 2013-01-01 05:00:00 JFK BQN N804JB B6 <NA> <NA>
5 2013 2013-01-01 06:00:00 LGA ATL N668DN DL <NA> <NA>
6 2013 2013-01-01 05:00:00 EWR ORD N39463 UA <NA> <NA>
7 2013 2013-01-01 06:00:00 EWR FLL N516JB B6 <NA> <NA>
8 2013 2013-01-01 06:00:00 LGA IAD N829AS EV <NA> <NA>
9 2013 2013-01-01 06:00:00 JFK MCO N593JB B6 <NA> <NA>
10 2013 2013-01-01 06:00:00 LGA ORD N3ALAA AA <NA> <NA>
# ℹ 336,766 more rows
# ℹ 5 more variables: model <chr>, engines <int>, seats <int>, speed <int>,
# engine <chr>
When we try to do this, however, we get a bunch of NAs. Why?
join_by(tailnum) is short for join_by(tailnum == tailnum) making these types of basic joins equi joins.
# A tibble: 336,776 × 14
year.x time_hour origin dest tailnum carrier year.y type
<int> <dttm> <chr> <chr> <chr> <chr> <int> <chr>
1 2013 2013-01-01 05:00:00 EWR IAH N14228 UA 1999 Fixed wing mu…
2 2013 2013-01-01 05:00:00 LGA IAH N24211 UA 1998 Fixed wing mu…
3 2013 2013-01-01 05:00:00 JFK MIA N619AA AA 1990 Fixed wing mu…
4 2013 2013-01-01 05:00:00 JFK BQN N804JB B6 2012 Fixed wing mu…
5 2013 2013-01-01 06:00:00 LGA ATL N668DN DL 1991 Fixed wing mu…
6 2013 2013-01-01 05:00:00 EWR ORD N39463 UA 2012 Fixed wing mu…
7 2013 2013-01-01 06:00:00 EWR FLL N516JB B6 2000 Fixed wing mu…
8 2013 2013-01-01 06:00:00 LGA IAD N829AS EV 1998 Fixed wing mu…
9 2013 2013-01-01 06:00:00 JFK MCO N593JB B6 2004 Fixed wing mu…
10 2013 2013-01-01 06:00:00 LGA ORD N3ALAA AA NA <NA>
# ℹ 336,766 more rows
# ℹ 6 more variables: manufacturer <chr>, model <chr>, engines <int>,
# seats <int>, speed <int>, engine <chr>If you have keys that have the same meaning (values) but are named different things in their respective datasets you’d also specify that with join_by()
by = c("dest" = "faa") was the former syntax for this and you still might see that in older code. You can specify multiple join_bys by simply separating the conditional statements with , (i.e. join_by(x == y, a == b)).
# A tibble: 336,776 × 13
year time_hour origin dest tailnum carrier name lat lon
<int> <dttm> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
1 2013 2013-01-01 05:00:00 EWR IAH N14228 UA George Bu… 30.0 -95.3
2 2013 2013-01-01 05:00:00 LGA IAH N24211 UA George Bu… 30.0 -95.3
3 2013 2013-01-01 05:00:00 JFK MIA N619AA AA Miami Intl 25.8 -80.3
4 2013 2013-01-01 05:00:00 JFK BQN N804JB B6 <NA> NA NA
5 2013 2013-01-01 06:00:00 LGA ATL N668DN DL Hartsfiel… 33.6 -84.4
6 2013 2013-01-01 05:00:00 EWR ORD N39463 UA Chicago O… 42.0 -87.9
7 2013 2013-01-01 06:00:00 EWR FLL N516JB B6 Fort Laud… 26.1 -80.2
8 2013 2013-01-01 06:00:00 LGA IAD N829AS EV Washingto… 38.9 -77.5
9 2013 2013-01-01 06:00:00 JFK MCO N593JB B6 Orlando I… 28.4 -81.3
10 2013 2013-01-01 06:00:00 LGA ORD N3ALAA AA Chicago O… 42.0 -87.9
# ℹ 336,766 more rows
# ℹ 4 more variables: alt <dbl>, tz <dbl>, dst <chr>, tzone <chr>You can request dplyr to keep both keys with keep = TRUE argument.
# A tibble: 336,776 × 14
year time_hour origin dest tailnum carrier faa name lat
<int> <dttm> <chr> <chr> <chr> <chr> <chr> <chr> <dbl>
1 2013 2013-01-01 05:00:00 EWR IAH N14228 UA IAH George Bu… 30.0
2 2013 2013-01-01 05:00:00 LGA IAH N24211 UA IAH George Bu… 30.0
3 2013 2013-01-01 05:00:00 JFK MIA N619AA AA MIA Miami Intl 25.8
4 2013 2013-01-01 05:00:00 JFK BQN N804JB B6 <NA> <NA> NA
5 2013 2013-01-01 06:00:00 LGA ATL N668DN DL ATL Hartsfiel… 33.6
6 2013 2013-01-01 05:00:00 EWR ORD N39463 UA ORD Chicago O… 42.0
7 2013 2013-01-01 06:00:00 EWR FLL N516JB B6 FLL Fort Laud… 26.1
8 2013 2013-01-01 06:00:00 LGA IAD N829AS EV IAD Washingto… 38.9
9 2013 2013-01-01 06:00:00 JFK MCO N593JB B6 MCO Orlando I… 28.4
10 2013 2013-01-01 06:00:00 LGA ORD N3ALAA AA ORD Chicago O… 42.0
# ℹ 336,766 more rows
# ℹ 5 more variables: lon <dbl>, alt <dbl>, tz <dbl>, dst <chr>, tzone <chr>right_join()Has the same interface as a left_join but keeps all rows in y instead of x
inner_join()Has the same interface as a left_join but only keeps rows that occur in both x and y
full_join()Has the same interface as a left_join but keeps all rows in either x or y
Filtering Joins

semi_join()Keeps all rows in x that have a match in y
semi_join() in nycflights13We could use a semi-join to filter the airports dataset to show just the origin airports.
# A tibble: 3 × 8
faa name lat lon alt tz dst tzone
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 EWR Newark Liberty Intl 40.7 -74.2 18 -5 A America/New_York
2 JFK John F Kennedy Intl 40.6 -73.8 13 -5 A America/New_York
3 LGA La Guardia 40.8 -73.9 22 -5 A America/New_Yorkanti_join()Returns all rows in x that don’t have a match in y
anti_join() in nycflights13We can find rows that are missing from airports by looking for flights that don’t have a matching destination airport.
# A tibble: 1,455 × 8
faa name lat lon alt tz dst tzone
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 04G Lansdowne Airport 41.1 -80.6 1044 -5 A America/…
2 06A Moton Field Municipal Airport 32.5 -85.7 264 -6 A America/…
3 06C Schaumburg Regional 42.0 -88.1 801 -6 A America/…
4 06N Randall Airport 41.4 -74.4 523 -5 A America/…
5 09J Jekyll Island Airport 31.1 -81.4 11 -5 A America/…
6 0A9 Elizabethton Municipal Airport 36.4 -82.2 1593 -5 A America/…
7 0G6 Williams County Airport 41.5 -84.5 730 -5 A America/…
8 0G7 Finger Lakes Regional Airport 42.9 -76.8 492 -5 A America/…
9 0P2 Shoestring Aviation Airfield 39.8 -76.6 1000 -5 U America/…
10 0S9 Jefferson County Intl 48.1 -123. 108 -8 A America/…
# ℹ 1,445 more rowsThere are three possible outcomes for a row in x:
y, it’s preserved.y, it’s duplicated once for each match.What happens if we match on more than one row?
Warning in inner_join(df1, df2, join_by(key)): Detected an unexpected many-to-many relationship between `x` and `y`.
ℹ Row 2 of `x` matches multiple rows in `y`.
ℹ Row 2 of `y` matches multiple rows in `x`.
ℹ If a many-to-many relationship is expected, set `relationship =
"many-to-many"` to silence this warning.# A tibble: 5 × 3
key val_x val_y
<dbl> <chr> <chr>
1 1 x1 y1
2 2 x2 y2
3 2 x2 y3
4 2 x3 y2
5 2 x3 y3 The joins we’ve discussed thus far have all been equi-joins, where the rows match if the x key equals the y key. But you can also specify other types of relationships.
dplyr has four different types of non-equi joins:
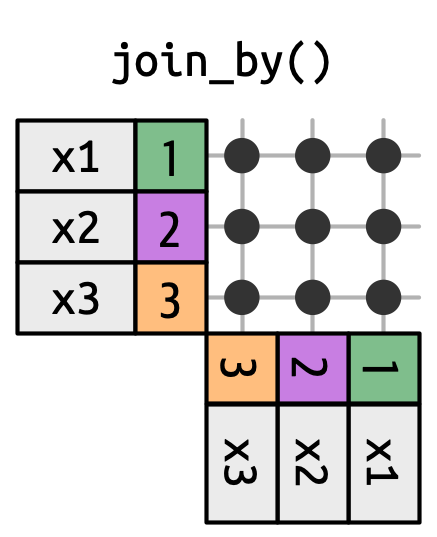
The joins we’ve discussed thus far have all been equi-joins, where the rows match if the x key equals the y key. But you can also specify other types of relationships.
dplyr has four different types of non-equi joins:
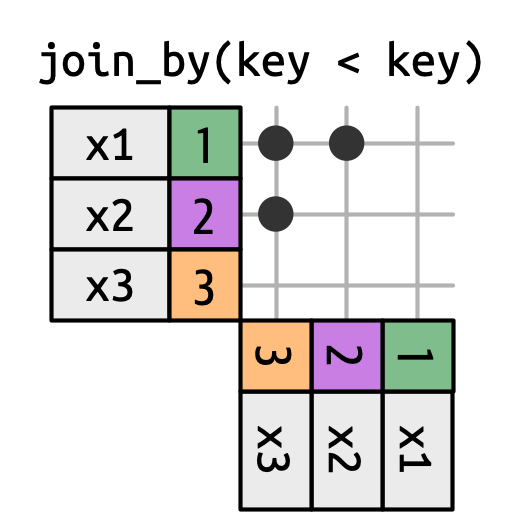
The joins we’ve discussed thus far have all been equi-joins, where the rows match if the x key equals the y key. But you can also specify other types of relationships.
dplyr has four different types of non-equi joins: